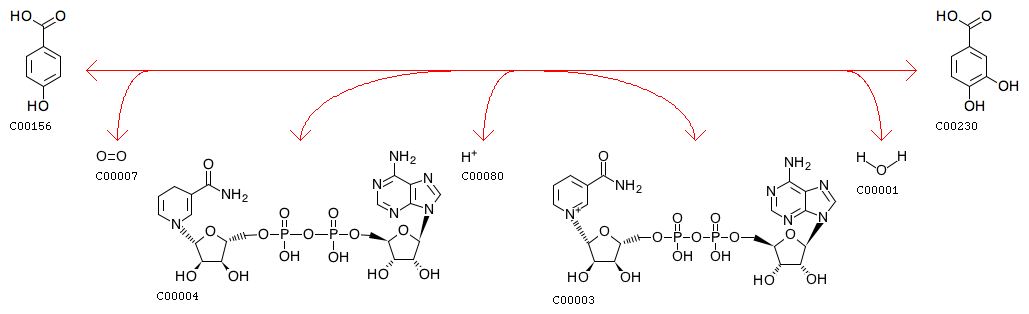
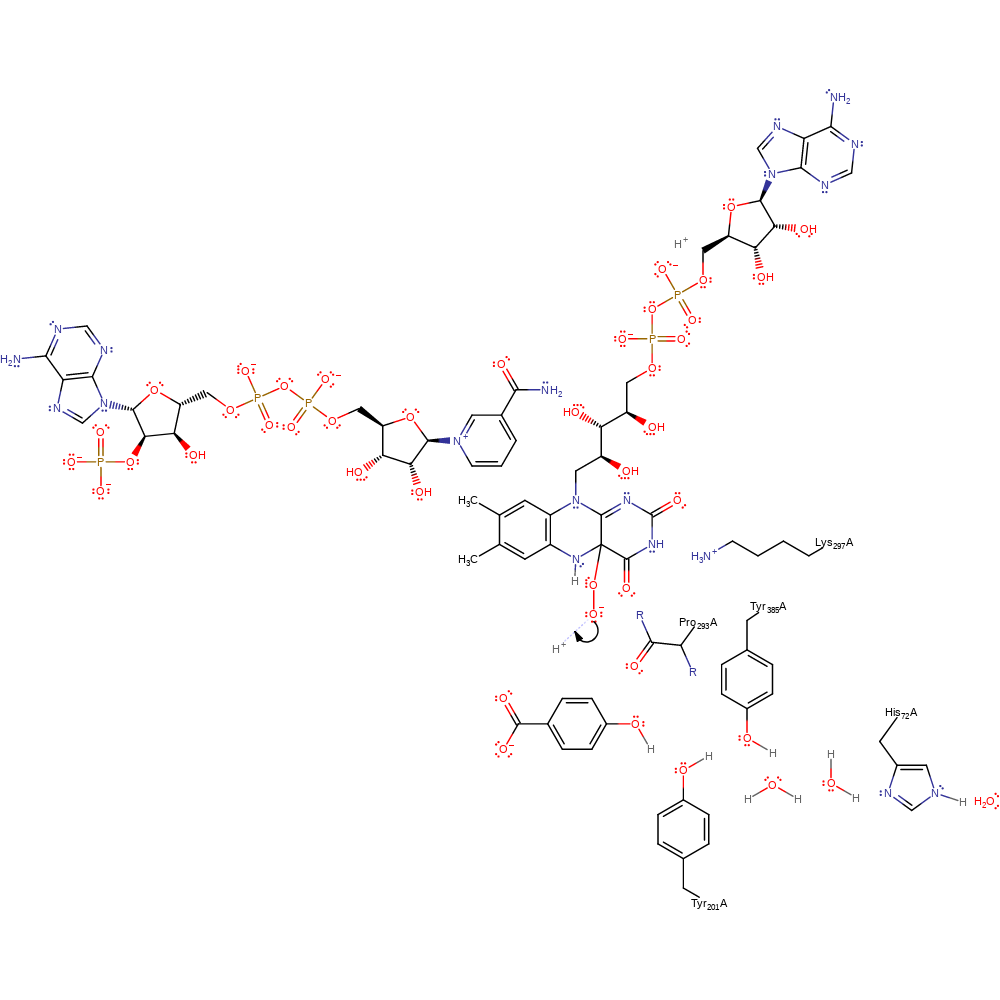
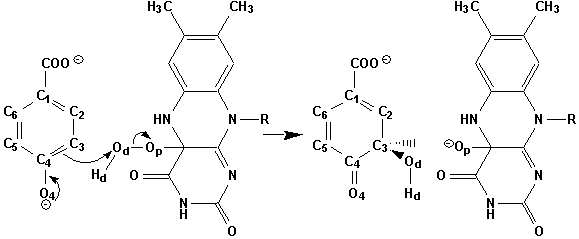
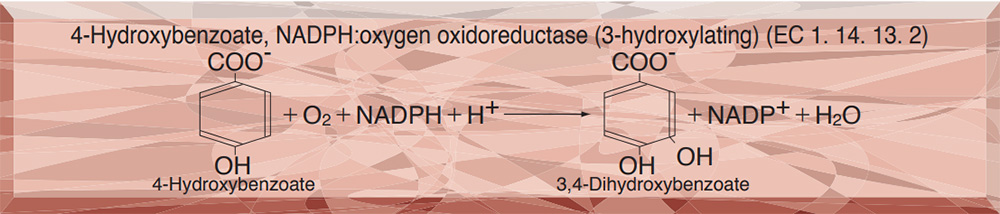
Oxygen Reactions in p-Hydroxybenzoate Hydroxylase Utilize the H-Bond Network during Catalysis | Biochemistry
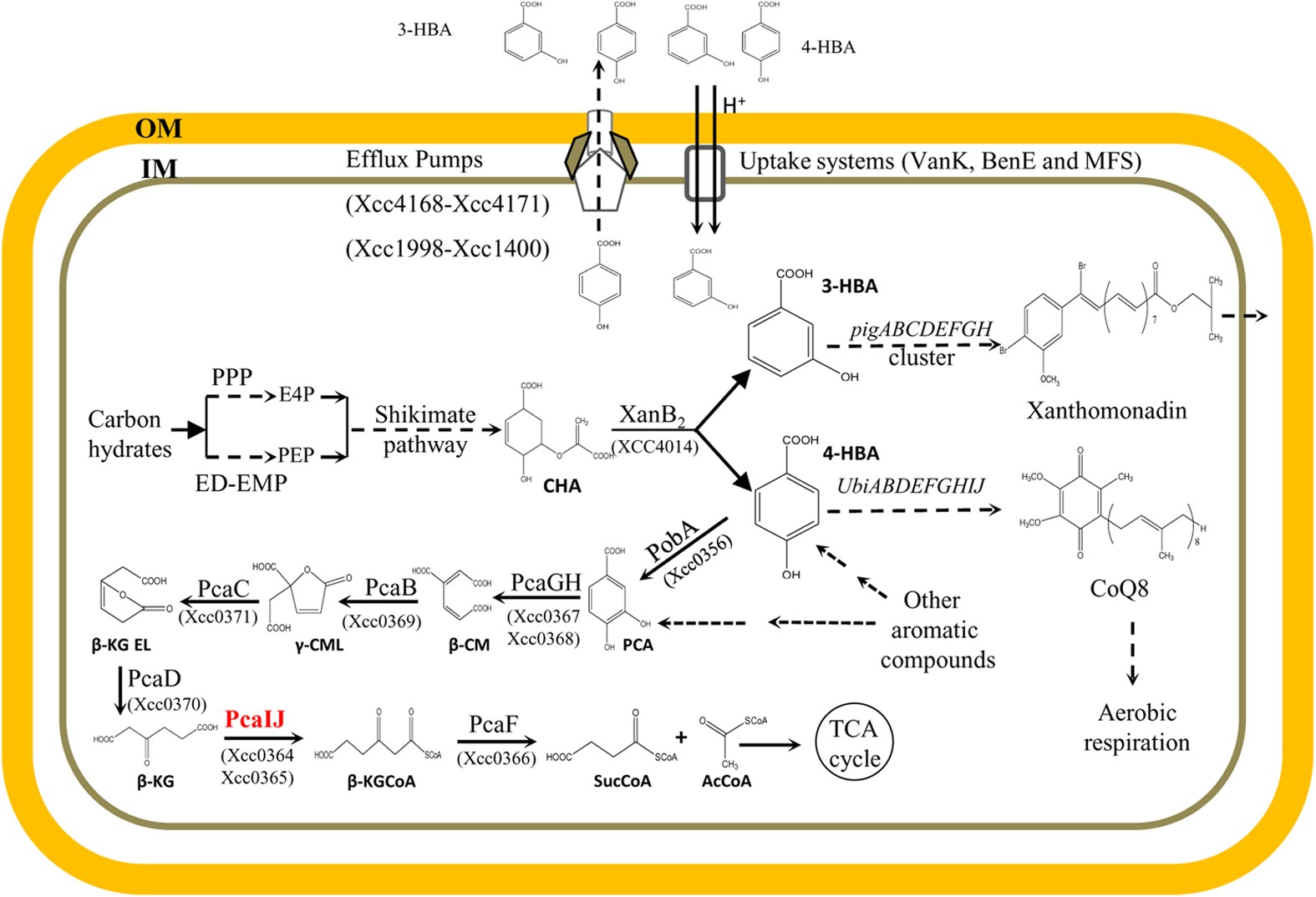
A functional 4-hydroxybenzoate degradation pathway in the phytopathogen Xanthomonas campestris is required for full pathogenicity | Scientific Reports
Genomic and Functional Analyses of the Gentisate and Protocatechuate Ring-Cleavage Pathways and Related 3-Hydroxybenzoate and 4-Hydroxybenzoate Peripheral Pathways in Burkholderia xenovorans LB400 | PLOS ONE
![PDF] Purification and Properties of 4-Hydroxybenzoate 1-Hydroxylase ( Decarboxylating ) , a Novel Flavin Adenine Dinucleotide-Dependent Monooxygenase from Candida parapsilosis CBS 604 | Semantic Scholar PDF] Purification and Properties of 4-Hydroxybenzoate 1-Hydroxylase ( Decarboxylating ) , a Novel Flavin Adenine Dinucleotide-Dependent Monooxygenase from Candida parapsilosis CBS 604 | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/fa660864696c850fc1b70c1a28724f636a2949c6/3-Figure1-1.png)
PDF] Purification and Properties of 4-Hydroxybenzoate 1-Hydroxylase ( Decarboxylating ) , a Novel Flavin Adenine Dinucleotide-Dependent Monooxygenase from Candida parapsilosis CBS 604 | Semantic Scholar
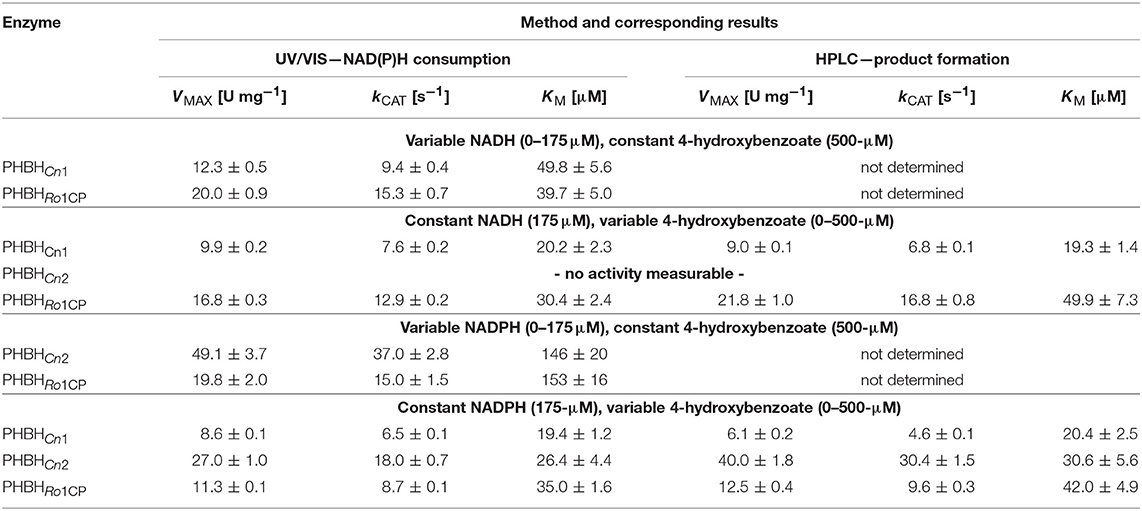
Frontiers | Pyridine Nucleotide Coenzyme Specificity of p-Hydroxybenzoate Hydroxylase and Related Flavoprotein Monooxygenases

Monooxygenation of aromatic compounds by flavin‐dependent monooxygenases - Chenprakhon - 2019 - Protein Science - Wiley Online Library

Understanding the Molecular Mechanism Underlying the High Catalytic Activity of p-Hydroxybenzoate Hydroxylase Mutants for Producing Gallic Acid | Biochemistry

Molecular and biochemical characterization of the xlnD-encoded 3- hydroxybenzoate 6-hydroxylase involved in the degradation of 2,5-xylenol via the gentisate pathway in Pseudomonas alcaligenes NCIMB 9867. - Abstract - Europe PMC

Altered Balance of Half-reactions in p-Hydroxybenzoate Hydroxylase Caused by Substituting the 2′-Carbon of FAD with Fluorine* - Journal of Biological Chemistry

RCSB PDB - 1BKW: p-Hydroxybenzoate hydroxylase (phbh) mutant with cys116 replaced by ser (c116s) and arg44 replaced by lys (r44k), in complex with fad and 4-hydroxybenzoic acid

A growth-based, high-throughput selection platform enables remodeling of 4-hydroxybenzoate hydroxylase active site | bioRxiv

ToF-SIMS imaging reveals that p-hydroxybenzoate groups specifically decorate the lignin of fibres in the xylem of poplar and willow
![PDF] The Structure of p-Hydroxybenzoate Hydroxylase by Herman Schreuder, Jan Metske Van Der Laan, Wim G. J. Hol, Jan Drenth · 10.1201/9781351070584-2 · OA.mg PDF] The Structure of p-Hydroxybenzoate Hydroxylase by Herman Schreuder, Jan Metske Van Der Laan, Wim G. J. Hol, Jan Drenth · 10.1201/9781351070584-2 · OA.mg](https://og.oa.mg/The%20Structure%20of%20p-Hydroxybenzoate%20Hydroxylase.png?author=%20Herman%20Schreuder,%20Jan%20Metske%20Van%20Der%20Laan,%20Wim%20G.%20J.%20Hol,%20Jan%20Drenth)
PDF] The Structure of p-Hydroxybenzoate Hydroxylase by Herman Schreuder, Jan Metske Van Der Laan, Wim G. J. Hol, Jan Drenth · 10.1201/9781351070584-2 · OA.mg
![PDF] Properties of p-hydroxybenzoate hydroxylase when stabilized in its open conformation. | Semantic Scholar PDF] Properties of p-hydroxybenzoate hydroxylase when stabilized in its open conformation. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/83e5e7248d4153cddfe890e9d9102b0836f867e7/2-Figure1-1.png)